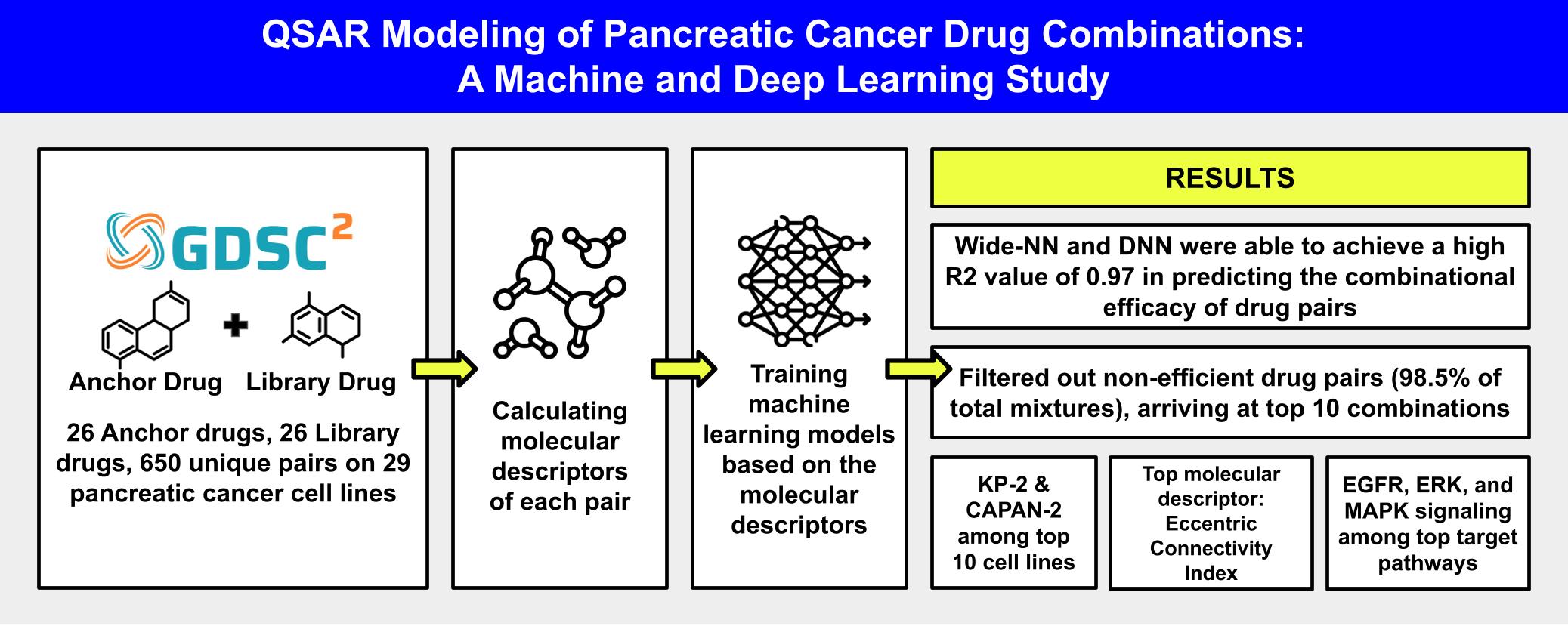
This study focuses on leveraging machine learning (ML) and deep learning (DL) algorithms to develop a structure-activity relationship between the molecular descriptors of drug pairs and their combined biological activity through a QSAR model. We utlized 11 ML/DL algorithms to create these QSAR models, using data from 29 pancreatic cancer cell lines, 26 anchor drugs, and 26 library drugs. Our results indicated that Wide Neural Network (Wide-NN) achieved a exceptional Coefficient of Determination (R²) score of 0.97 and a Root Mean Square Error (RMSE) of 0.51, making it the most effective algorithm for developing a robust structure-activity relationship with strong generalization capabilities. Combining therapy with ML and DL techniques for accelerating drug discovery offers a promising approach to tackling pancreatic cancer.
The original dataset utilized in this study was obtained from Genomics of Drug Sensitivity in Cancer (GDSC2) combinations database. You can directly download the original dataset from here. The original dataset is also available in this repository via pancreas_anchor_combo.csv.
The original pancreatic cancer GDSC2 combinational dataset does not contain SMILES strings for the anchor and library drugs. Therefore, we have calculated the SMILES for the mentioned molecules with the help of PubChemPy python library. We then calculated the molecular descriptors for each unique molecule with PadelPy. The calculated descriptors for anchor and library drugs are stored in anchor_descriptors.csv and library_descriptors.csv files.
After calculating the molecular descriptors, we then concatenated the descriptors files with the original dataset, extending it. The final dataset ready to go through pre-processing and model training is available from this Google Drive link The size of the dataset is close to 3 gigabytes.
Everything that is needed to replicate our study can be found in pancreatic_cancer_combinational_ml.ipynb jupyter notebook. The code for EDA, SMILES convertion, molecular descriptors calculation, pre-processing, model training, and model interpretation is accesible within the mentioned file.