Releases: data-others/brain
NODDI Lifespan dMRI (Chang & Mukherjee, 2015)
Diffusion MRI from 67 healthy controls (ages 7–63 years) to study white-matter maturation with NODDI (neurite orientation dispersion and density imaging), alongside standard DTI metrics. The release includes multi-shell HARDI (two shells), single-shell DTI, and T1-weighted anatomical images suitable for ROI- and voxel-wise analyses of neurite density (ND), orientation dispersion (OD), and conventional diffusion indices. Subsets include test-retest sessions for reliability assessment. Findings reported in the companion article show ND rising with a logarithmic pattern across childhood to adulthood, OD increasing exponentially later in life, and NODDI measures predicting age more specifically than DTI.
License
Use is governed by the dataset’s UCSF Datashare Data Use Agreement (see Dryad record).
Citation
Chang, Y., & Mukherjee, P. (2015). NODDI-PLOS-ONE-Chang et al 2015 [Data set]. Dryad. https://doi.org/10.7272/Q6D798BD
Primary article: PLOS ONE — Neurite orientation dispersion and density imaging of brain development across the lifespan (2015).
Source
https://doi.org/10.7272/Q6D798BD
Repository: Dryad (Published Mar 2, 2015)
Dataset details
- Participants: 67 healthy controls, 7–63 years; includes test-retest subsets.
- Modalities:
- DTI: single-shell diffusion (b≈1000 s/mm²).
- HARDI (×2): multi-shell diffusion for NODDI modeling.
- Anatomical: T1-weighted MRI.
- Intended uses: lifespan modeling of ND/OD, age prediction, reliability benchmarking, comparison of NODDI vs. DTI.
Suggested acknowledgement
Please cite the Dryad dataset DOI above and the associated PLOS ONE article when using these data.
Keywords
NODDI • Diffusion MRI • DTI • Lifespan • White matter • Development • Test–retest
DTI Predicts Mandarin Learning
This dataset contains diffusion tensor imaging (DTI) data and corresponding behavioral learning measures from a study examining how white matter microstructure predicts success in learning Mandarin Chinese as a second language.
Each subject’s dataset includes nifti-formatted DWI volumes, b-values, and b-vectors, along with a separate CSV file containing demographic and learning performance information.
The dataset accompanies the publication:
Qi Z., Han M., Garel K., Chen E. S., & Gabrieli J. D. E. (2014). White-matter structure in the right hemisphere predicts Mandarin Chinese learning success. Journal of Neurolinguistics, 33, 14–28.
License
Creative Commons Attribution 4.0 International (CC BY 4.0)
Citation
Qi, Z. (2017).
DTI Predicts Mandarin Learning [Data set]. Zenodo.
https://doi.org/10.5281/zenodo.260007
Source
https://doi.org/10.5281/zenodo.260007
Contact: zhenghan.qi@unl.edu
Institution: University of Nebraska–Lincoln, Department of Special Education and Communication Disorders
Dataset Information
| Category | Details |
|---|---|
| Subjects | De-identified adult participants learning Mandarin as a second language |
| Study Type | Diffusion MRI study investigating neuroanatomical predictors of second language learning |
| Data Format | NIfTI for diffusion volumes, .bval and .bvec text files, and a .csv file with demographic and behavioral data |
| Imaging Modality | Diffusion Tensor Imaging (DTI) |
| Behavioral Measures | Mandarin learning success scores and demographic metadata |
| Anonymization | All participant identifiers removed prior to data sharing |
| Analysis Context | Investigates the relationship between right-hemisphere white matter integrity and language acquisition success |
Purpose
This dataset supports research into the neurobiological basis of second language learning, with an emphasis on white matter connectivity and diffusion anisotropy as predictors of individual learning outcomes.
It is useful for validating DTI-based biomarkers of language aptitude and for developing new analytic frameworks linking microstructural brain features to language learning performance.
Keywords
Diffusion MRI • DTI • Language Learning • Mandarin Chinese • Second Language Acquisition • White Matter • Neuroplasticity • Neurolinguistics • FA • Tractography
Southwest University Longitudinal Imaging Multimodal (SLIM) Brain Data Repository: A Long-term Test-Retest Sample of Young Healthy Adults in Southwest China
The SLIM (Structural, Longitudinal, and Integrated Multimodal) dataset is a valuable resource in neuroimaging, acquired through multimodal magnetic resonance imaging (mMRI). With a three-and-a-half-year longitudinal design, it addresses gaps in test-retest reliability and age-span limitations. Featuring diverse mMRI scans and behavioral assessments, SLIM provides a comprehensive view of the human brain. This dataset, acquired meticulously, is available for researchers globally, fostering collaboration and contributing to reproducible human brain sciences in partnership with CoRR. Explore the SLIM dataset for insights into the complexities of the human brain.
License
License: Creative Commons License: Attribution - Non-Commercial
Reference: Liu W, Wei D, Chen Q, Yang W, Meng J, Wu G, Bi T, Zhang Q, Zuo XN, Qiu J. Longitudinal test-retest neuroimaging data from healthy young adults in southwest China. Scientific data. 2017 Feb 14;4(1):1-9. link
Official website: link
Download
| File Format | Modality/Content | Link | Details |
|---|---|---|---|
| SRC | Eddy corrected DWI | OneDrive | The SRC files contain raw dMRI signals for modeling. They can be reconstructed in DSI Studio to generate FIB files. The original NIFTI files can be downloaded from the HCP's connectome db website. |
| FIB | Fiber orientation maps in the native space | OneDrive | GQI reconstructed FIB file in the native space. The FIB files are track-ready files for DSI Studio to run fiber tracking. |
| NIFTI | T1W | OneDrive | |
| Text file | demographics | OneDrive |
Methods
A DTI diffusion scheme was used, and a total of 90 diffusion sampling directions were acquired. The b-value was 1000 s/mm². The in-plane resolution was 2 mm. The slice thickness was 2 mm. FSL eddy was used to correct for eddy current distortion. The correction was conducted through the integrated interface in DSI Studio ("Chen" release)(http://dsi-studio.labsolver.org). The diffusion MRI data were rotated to align with the AC-PC line at an isotropic resolution of 2.000000. The restricted diffusion was quantified using restricted diffusion imaging (Yeh et al., MRM, 77:603–612 (2017)). The diffusion data were reconstructed using generalized q-sampling imaging (Yeh et al., IEEE TMI, ;29(9):1626-35, 2010) with a diffusion sampling length ratio of 1.25. The tensor metrics were calculated using DWI with b-value lower than 1750 s/mm².
Stark Cross-Sectional Aging Dataset
This dataset includes behavioral and neuroimaging data from approximately 120 participants aged 18 to 89 years, collected as part of a research grant focused on understanding the effects of aging on the hippocampus and its relationship to age-related cognitive decline.
The project combines high-resolution imaging techniques with advanced behavioral assessments to provide insights into structural and functional brain changes across the lifespan.
Note: Some, but not all, of the data are currently live. Additional data will be uploaded as time and storage capacity allow.
Available Data
https://www.nitrc.org/projects/stark_aging/
-
Traditional Neuropsychological Assessments
-
Hippocampal-Specific Behavioral Tasks
-
Whole-Brain Diffusion Tensor Imaging (DTI)
-
High-Resolution DTI of the Medial Temporal Lobes
-
Structural MRI Data:
- Segmentation of gray matter, white matter, and cerebrospinal fluid (CSF)
- Cortical parcellations using FreeSurfer
- Segmentation of hippocampal subfields
Download and Access
- Whole-Brain DTI Data:
Download DTI_WB_75-89.tar.bz2 (Approx. 340 MB)
Additional data will become available as it is processed and uploaded.
License
This dataset is distributed under a Creative Commons Attribution-NonCommercial-ShareAlike License (CC BY-NC-SA).
Penthera 3T
The acquisitions were done by the Sherbrooke Connectivity Imaging Lab in 2016. Thirteen young and healthy subjects (three women and ten men, mean of 25.92 (+-1.85) years old) were scanned six times: two sessions with three scans per session. The DWI (112x112 matrix, TR 5615 ms, TE 95 ms, SENSE factor of 2) were acquired on a 3 Tesla MRI (Philips, Ingenia) with a single-shot echo-planar imaging sequence having 3 different shells, b=300, 1000, 2000 mm2/s with respectively 8, 32 and 60 directions distributed, 7 b=0 mm2/s for a total of 107 images and a 2 mm isotropic spatial resolution. A reversed phase encoded b=0 image is available, to correct EPI distortions, with the same spatial resolution as the DWI. The T1-weighted MPRAGE image (TR 7.9 ms, TE 3.5 ms) has a 1mm isotropic spatial resolution.
Source
https://doi.org/10.5281/zenodo.2602049
Paquette, Michael1
Gilbert, Guillaume2
Descoteaux, Maxime1
NTU 90
This repository hosts the NTU‑90 diffusion MRI dataset, comprising 90 subjects scanned on a Siemens 3 T Trio using a diffusion spectrum imaging (DSI) protocol (203 directions, b‑max=6000 s/mm²) with 3 mm isotropic resolution and a one‑subject 2 mm DSI scan (b‑max=4000 s/mm²). Data have been converted to NIfTI for broad compatibility.
License
The NTU Lab grants a non‑commercial, educational, and research license for use of these images with the following conditions:
- Always acknowledge the Advanced Biomedical MRI Lab, NTU Hospital in any publications or materials.
- Redistribution is permitted only if this paragraph is included. Users assume all risk and liability.
- NTU Lab provides no warranties of any kind, express or implied.
- Commercial use is strictly prohibited without written consent from NTU Lab.
Data Acquisition
3 mm DSI (n=90)
- Scanner: Siemens Trio 3 T (8‑channel head coil)
- Sequence: Twice‑refocused spin‑echo EPI
- TE/TR: 142 ms / 9 100 ms
- Directions: 203 on a Cartesian grid, b‑max=6 000 s/mm²
- Voxel size: 2.9 × 2.9 × 2.9 mm³
- Scan time: ~45 min per subject
References
- Yeh, F.-C., & Tseng, W.-Y.I. NTU-90: a high angular resolution brain atlas constructed by q-space diffeomorphic reconstruction. NeuroImage 58(1), 91–99 (2011).
- Yeh, F.-C., Wedeen, V.J., & Tseng, W.-Y.I. Generalized q-sampling imaging. IEEE Trans. Med. Imaging 29(9), 1626–1635 (2010).
The NKI Rockland Sample
The NKI Rockland Sample Initiative, funded by the US National Institutes of Health and NY State Office of Mental Health, is a cutting-edge research program mapping the human brain's development and its connections to behavior. With over 1,500 participants from Rockland County and nearby areas, this initiative actively involves individuals of all ages in shaping our understanding of mental health. For researchers, the NKI-Rockland Sample offers a well-supported, rich neuroimaging and phenotypic resource, fostering collaborative efforts to advance knowledge on normative brain-behavior relationships across the lifespan.
License
NKI-Rockland Sample data are distributed using the Creative Commons-Attribution-Noncommercial license. For the high-dimensional phenotypic data, all terms specified by the DUA must be complied with.
Participants and Experiments
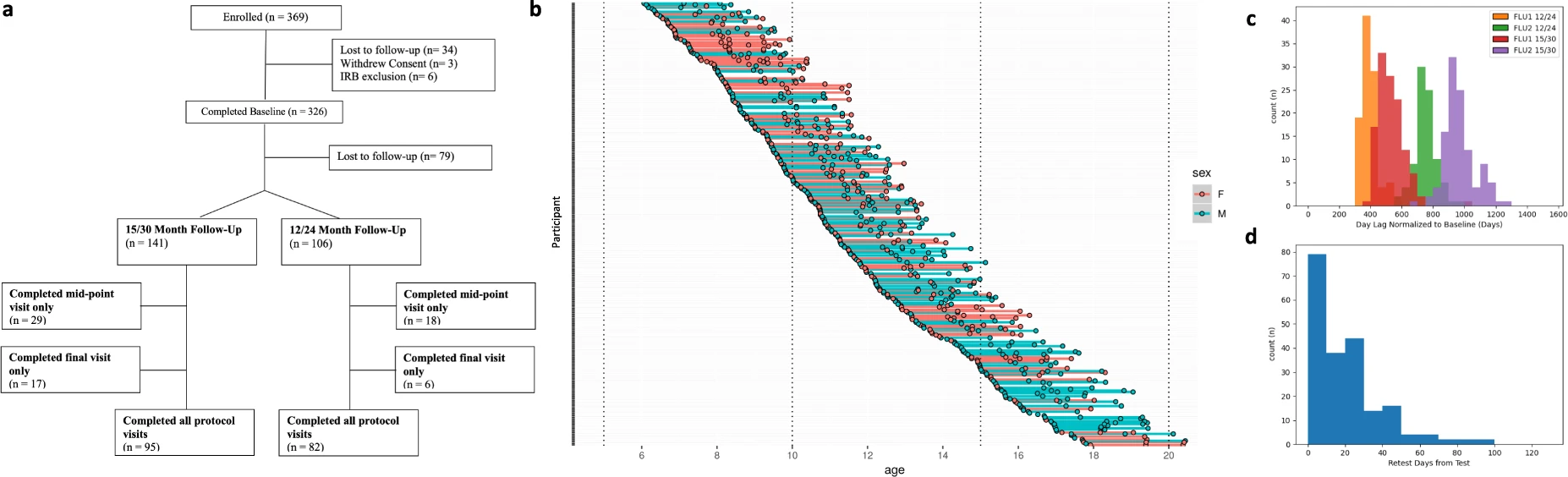
The enhanced Nathan Kline Institute-Rockland Sample I was an institutionally centered endeavor aimed at creating a large-scale community neuroimaging sample of participants across the lifespan (ages 6-85 years). Measures included a wide array of physiological and behavioral assessments, cognitive and psychiatric characterization, genetic information, and advanced neuroimaging. Anonymized data are publicly shared openly and were released prospectively (i.e., on a quarterly basis) as data were collected. The NKI Rockland Sample I data resource includes N = 1, 500 participants across the first set of NIH funded studies (2011-2020).
Phenotypic data
Phenotypic data can be accessed at COINS Data Exchange. Except for age, sex, and handedness, which are publicly available, NKI-RS phenotypic data are protected by a Data Usage Agreement (DUA). Investigators must complete the DUA and have it approved by an authorized institutional official before receiving access.
The NKI-Rockland phenotypic battery primarily focuses on dimensional mental health assessments, particularly psychiatric symptomatology detection (e.g., Conners ADHD Scale, Children's Depression Inventory). However, the reliance on deficit-only measures, common in the field, is questioned for its limitations in distinguishing behaviors among unaffected individuals. Recent efforts highlight this concern, emphasizing the need for measures assessing strengths as well as weaknesses. For ADHD assessment, both Conners (symptoms) and SWAN (strengths and weaknesses) are included, enabling exploration of this issue. While attempts were made to include measures emphasizing strength, standard clinical measures remain central due to the lack of assessments in psychiatric symptom characterization. The recommendation is to incorporate psychometric instruments producing dimensional distributions in non-clinical samples for community sample phenotyping.
-
BAS1: BASELINE1
-
BAS2: BASELINE2
-
FLU1: FOLLOWUP1 (mid-point follow-up)
-
FLU2: FOLLOWUP2 (final follow-up)
-
TRT: RETEST BAS1 (usually acquired within 3 weeks of a baseline or a follow-up)
Multi-Modal MRI Reproducibility Resource
This repository hosts the **Multi‑Modal MRI Reproducibility Resource **, a publicly available collection of scan–rescan MRI sessions on 21 healthy volunteers acquired on a 3 T scanner using a standardized one‑hour multi‑parametric protocol. It comprises structural (MPRAGE, FLAIR), diffusion (DTI), functional (resting‑state fMRI), field‑mapping (B0/B1), perfusion (ASL, VASO), quantitative relaxometry (T1, T2), and magnetization transfer imaging, all converted to NIfTI format. This dataset is intended for statisticians and imaging scientists to benchmark and quantify reproducibility across diverse MRI modalities.
Overview
The Multi‑Modal MRI Reproducibility Resource provides scan–rescan imaging sessions of 21 healthy volunteers (no history of neurological disease) acquired within a clinically feasible 60‑minute protocol on a 3 T MRI scanner. It serves as a gold standard for evaluating reproducibility and power analyses in structural, functional, and microstructural imaging methods.
Imaging Protocol
- Structural: MPRAGE, FLAIR
- Diffusion: DTI (30+ directions)
- Functional: Resting‑state fMRI
- Field Maps: B0 and B1 acquisitions
- Perfusion: ASL, VASO
- Relaxometry & MT: Quantitative T1, T2, and magnetization transfer imaging
All data were originally in Philips PAR/REC format and have been converted to NIfTI for broad compatibility.
This structure facilitates targeted analysis pipelines for each modality.
Citation
When using this dataset, please cite:
Bennett A. Landman, Alan J. Huang, Aliya Gifford, Deepti S. Vikram, Issel A. L. Lim, Jonathan A. D. Farrell, John A. Bogovic, Jun Hua, Min Chen, Samson Jarso, Seth A. Smith, Suresh Joel, Susumu Mori, James J. Pekar, Peter B. Barker, Jerry L. Prince, and Peter C. M. van Zijl.
Multi‑Parametric Neuroimaging Reproducibility: A 3 T Resource Study. NeuroImage, 2010. NIHMS/PMC:252138. doi:10.1016/j.neuroimage.2010.11.047
License
Data are distributed under the BIRN Data License; acknowledgement of the original publication is requested upon reuse. Refer to the NITRC project page and the original publication for full license details.
https://www.nitrc.org/projects/multimodal/
Acknowledgements
Funded by NIH Grant 5R24EB029173 as part of the Biomedical Informatics Research Network (BIRN) initiative.
IXI Dataset (Institute of Psychiatry)
The IXI Dataset provides nearly 600 MRI scans from normal, healthy subjects, collected as part of the Information eXtraction from Images (IXI) project (EPSRC GR/S21533/02).
Dataset Description
Each subject underwent a standardized MRI protocol including:
- T1-weighted images
- T2-weighted images
- PD-weighted images
- Magnetic Resonance Angiography (MRA)
- Diffusion-weighted images (15 directions)
MRI data were collected at three hospitals in London:
| Hospital | Scanner | Field Strength |
|---|---|---|
| Hammersmith Hospital | Philips | 3T |
| Guy’s Hospital | Philips | 1.5T |
| Institute of Psychiatry | GE | 1.5T |
Download Links
All images are provided in NIfTI (.nii.gz) format and can be downloaded directly from the IXI project site:
- T1 images
- T2 images
- PD images
- MRA images
- DTI images (includes
bvecs.txtandbvals.txt) - Demographic information (spreadsheet)
License
This dataset is distributed under the Creative Commons Attribution-ShareAlike 3.0 International (CC BY-SA 3.0) license.
You are free to share and adapt the material, provided that appropriate credit is given and any derivatives are shared under the same license.
Please acknowledge the source of the IXI data, e.g.:
“Data provided by the IXI project, Imperial College London (https://brain-development.org/ixi-dataset/).”
Citation
If you use this dataset, please cite the IXI project and acknowledge support from:
Biomedical Image Analysis Group, Imperial College London
South Kensington Campus, London SW7 2AZ
Tel: +44 (0)20 7589 5111
IXI Dataset (Hammersmith Hospital)
The IXI Dataset provides nearly 600 MRI scans from normal, healthy subjects, collected as part of the Information eXtraction from Images (IXI) project (EPSRC GR/S21533/02).
Dataset Description
Each subject underwent a standardized MRI protocol including:
- T1-weighted images
- T2-weighted images
- PD-weighted images
- Magnetic Resonance Angiography (MRA)
- Diffusion-weighted images (15 directions)
MRI data were collected at three hospitals in London:
| Hospital | Scanner | Field Strength |
|---|---|---|
| Hammersmith Hospital | Philips | 3T |
| Guy’s Hospital | Philips | 1.5T |
| Institute of Psychiatry | GE | 1.5T |
Download Links
All images are provided in NIfTI (.nii.gz) format and can be downloaded directly from the IXI project site:
- T1 images
- T2 images
- PD images
- MRA images
- DTI images (includes
bvecs.txtandbvals.txt) - Demographic information (spreadsheet)
License
This dataset is distributed under the Creative Commons Attribution-ShareAlike 3.0 International (CC BY-SA 3.0) license.
You are free to share and adapt the material, provided that appropriate credit is given and any derivatives are shared under the same license.
Please acknowledge the source of the IXI data, e.g.:
“Data provided by the IXI project, Imperial College London (https://brain-development.org/ixi-dataset/).”
Citation
If you use this dataset, please cite the IXI project and acknowledge support from:
Biomedical Image Analysis Group, Imperial College London
South Kensington Campus, London SW7 2AZ
Tel: +44 (0)20 7589 5111