Documentation | Install | Tutorials | Forum
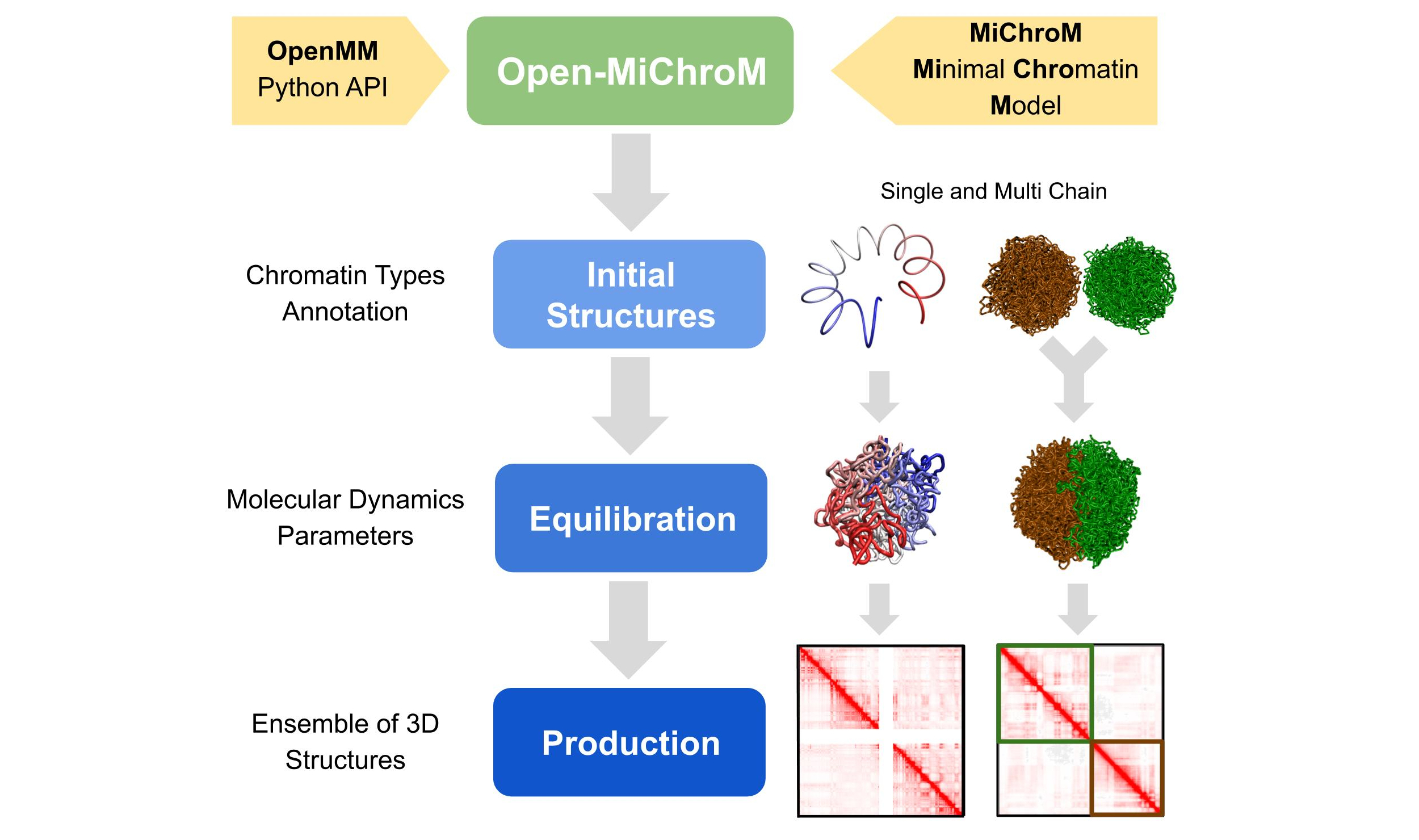
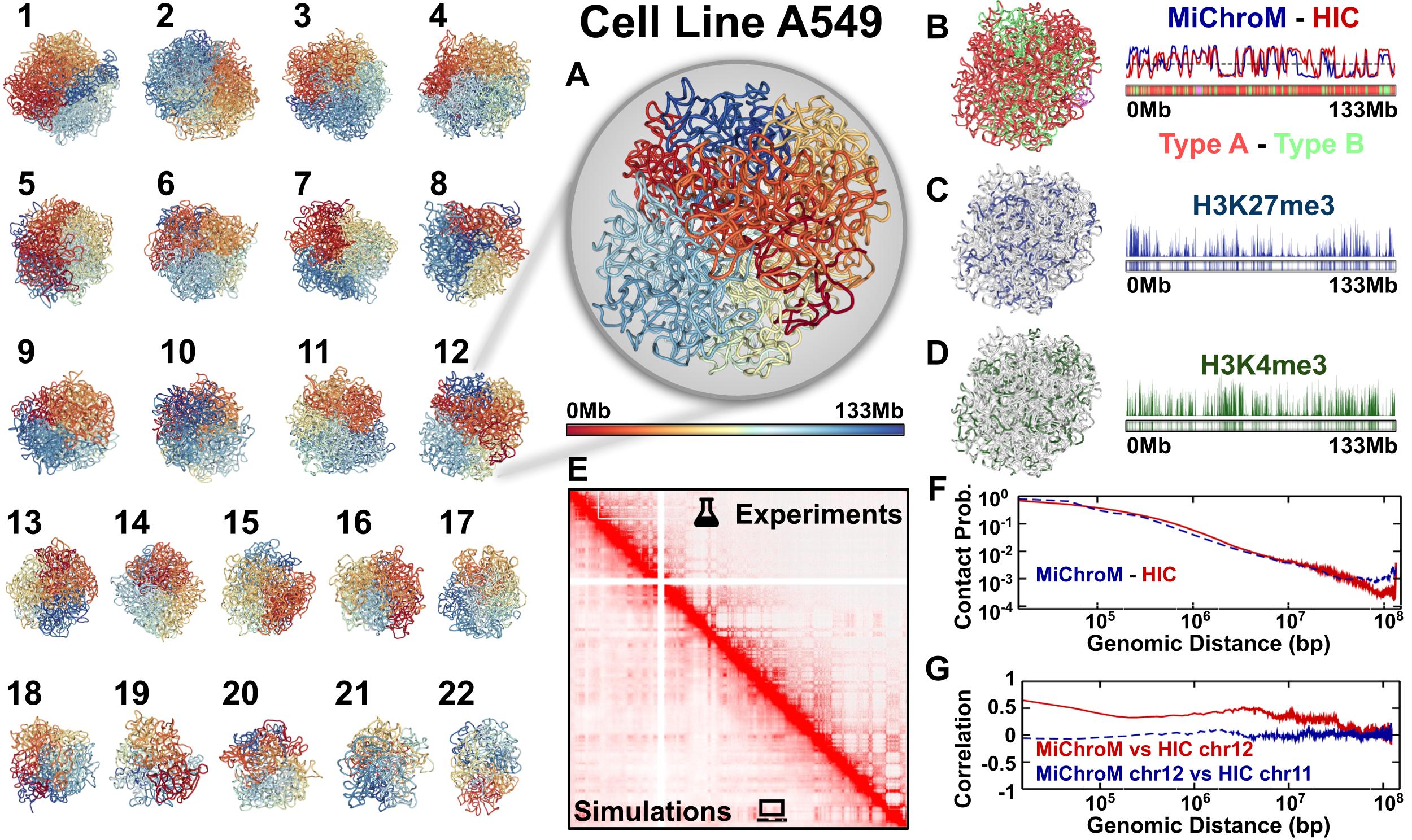
OpenMiChroM is a Python library for performing chromatin dynamics simulations and analyses. OpenMiChroM uses the OpenMM Python API employing the MiChroM (Minimal Chromatin Model) energy function. The chromatin dynamics simulations generate an ensemble of 3D chromosomal structures that are consistent with experimental Hi-C maps. OpenMiChroM also allows simulations of a single or multiple chromosome chains using high-performance computing on different platforms (GPUs and CPUs). It is a highly flexible framework that can be extended for chromatin modeling and simulations across different species and for general biomolecular simulations.
The chromatin dynamics simulations can be performed for different human cell lines, cell phases (interphase to metaphase), and various organisms from DNAzoo. Chromatin subcompartment annotations are available at the NDB (Nucleome Data Bank). The OpenMiChroM package accepts the chromatin sequence of compartments and subcompartments as input to create and simulate a chromosome polymer model. Examples of running the simulations and generating in silico Hi-C maps can be found here.
The following code snippet shows how to generate a single chromosome polymer model and run a chromatin dynamics simulation using OpenMiChroM:
from OpenMiChroM.ChromDynamics import MiChroM
sim = MiChroM(name='stomach_GRCh38', temperature=1.0, timeStep=0.01)
sim.setup(platform="cuda")
sim.saveFolder('stomach_GRCh38_chr10_simulation')
sim.buildClassicMichrom(ChromSeq='inputs/stomach_GRCh38.bed', chromosome='chr10')
sim.createReporters(statistics=True, traj=True, outputName=None, trajFormat="cndb", energyComponents=True, interval=10**3)
sim.run(nsteps=10**5, report=True, interval=10**4)
- Reference Documentation: Examples, tutorials, and class details.
- Installation Guide: Instructions for installing OpenMiChroM.
- GitHub repository: Download the OpenMiChroM source code.
- Issue tracker: Report issues/bugs or request features.
When using OpenMiChroM for chromatin dynamics simulations or analyses, please use this citation. We also thank Polychrom, where part of this code was inspired. You can use this citation.
The OpenMiChroM library can be installed via pip, conda, or compiled from source.
The code below will install OpenMiChroM from PyPI:
pip install OpenMiChroM
Note
OpenMiChroM relies on the OpenMM API to run the chromatin dynamics simulations.
OpenMM is now available as a pip-installable package. You can install it using pip openmm[cuda12] to iinstall to use with GPU's or openmm to install for CPU's only:
pip install openmm[cuda12]Alternatively, if you prefer to use conda, install OpenMM from the conda-forge channel with:
conda install -c conda-forge openmmIf you prefer using conda, you can install OpenMiChroM from conda-forge with the following command:
conda install -c conda-forge OpenMiChroM
Hint
Sometimes, the installation via conda may appear to be stuck. If this happens, update conda/anaconda using the command below and try installing OpenMiChroM again.
conda update --prefix /path/to/anaconda3/ anaconda
The following libraries are required for installing OpenMiChroM: